What is the
Simulaton Life?
The simulaton life is a rich life experience provided by training our
minds to consider simulations of natural and human phenomena often
in order to gain depth in understanding, awareness, and compassion.
From the Book
Book text is in a living document format, licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. In other words, you are free to share this work for non-commercial purposes as long as you don't create derivative works of this material, attribute me as the author, and keep this license notice intact. For more information about this Creative Commons license, please visit: http://CreativeCommons.org.
Chapter 8
biotic evolution simulation case study
Our second case study looks at simulation approaches to simulate evolutionary biology in plants. The theory of how plants evolve through genetic processes is simulated quite readily via a variety of approaches. Growth models do a satisfactory job of simulating a plant growing as it expresses its genetic information. The simulations provide our brains the opportunity to develop insights and deepen our understanding of evolutionary process in general.
Before we get into the details of the technical simulation tools available for use in this chapter, let's take a step back to see where they have come from historically. Gregor Mendel first solved the mystery of inheritance by experimenting with pea plants in the 19th century, leading to the eventual addition of the words allele, dominant, recessive, and diploid to common English vocabulary. Gregor Mendel started his life by following his ancestors in maintaining a family farm through gardening and beekeeping at a young age. After a formal university education and study for the priesthood, he lived at St Thomas's Abbey in St. Brno where he worked at plant breeding — watching the process across generations of plants closely. With the patience required of watching plants grow generation after generation between 1856 and 1863, he cultivated and tested some 29,000 pea plants and reported that one in four pea plants had purebred recessive alleles, two out of four were hybrid and one out of four were purebred dominant. His experiments led him to make two generalizations, the Law of Segregation and the Law of Independent Assortment, which later became known as Mendel's Laws of Inheritance.
Because the mathematics aligned well for such a large sample size of plants, those who read his published papers had the opportunity to imagine what possible vehicle in a pea plant would pass on that potential expression in future generations. But, sadly, Mendel's work was rejected at first and not widely accepted until after he died — the evidence was too revolutionary for those who clung to existing theories to accept. It would be ninety years after Mandel finished his pea plant experiments until the discovery of the structure of DNA in 1953. By then, a strong healthy competition was underway to find the physical structures in plants that could explain genetic expression over generations. Documentaries that track the work Watson and Crick performed to find the structure of DNA show the power that physical artifacts provided in helping arrive at strong candidates for DNA structure. Today, Watson and Crick would have had a tremendous number of virtual artifact approaches they could have relied on to help them discover DNA structure. These virtual artifacts are helping humanity study protein folding and other structures critical to plant development and health.
While Mendel started a trajectory of research into plant genetics, Aristid Lindenmayer started a trajectory of research into plant growth and development with his work with yeast and filamentous fungi and studies of growth patterns of various types of algae, such as blue-green bacteria. Generalizations of the growth process led Lindenmayer to develop a type of formal language called L-systems (or Lindenmayer Systems) in 1968. L-systems were devised to provide a formal description of the development of simple multi-cellular organisms, and to illustrate the neighborhood relationships between plant cells. From a basic starting base, L-systems have been extended to describe higher plants and complex branching structures associated with their growth.
Together Mendel and Lindenmayer initiated paths towards human understanding of how plants store information and then use those stored instructions to express their form during growth. Biological modeling and visualization researchers like Dr. Przemyslaw Prusinkiewicz at the University of Calgary have been working to apply understanding towards the generation of graphics that represent plants faithfully. As a case study in this chapter, we'll play with some software that let's us get a sense of what biological modelers do with software that can simulate plant growth over generations. We can then discuss possible directions to take the software for those of us who want to explore specific aspects of the simulation such as the work others have done previously.
In 1992, Deborah Fowler investigated the use of collisions while implementing the process of Spiral Phyllotaxis as a guideline to creating virtual plant models. Phyllotaxis is a well-documented phenomenon in plants that determines where primordia (seeds, petals, nuggets etc.) are located on a receptacle (sunflower, coneflower, corn ear) using a fixed divergence angle of 137.5 degrees. Primordia are packed following the rules of phyllotaxis until they collide with each other. Phyllotaxis has been incorporated into all plant development simulation systems as a result of Fowler and others' work. In 1996, Radomir Mech devised a system design where virtual plants could be modularly developed through interaction with other environmental modules. Using such a system, virtual plants don' t grow in a vacuum, but instead have their growth parameters affected by dynamic environmental parameters. For example, if the environment experiences a drought, plant growth is stunted and body structures wilt. The communication goes in both directions. If a species of plant becomes successful in growing and respiring, additional oxygen is passed back to the environment. In 1998, Oliver Deussen led a team that focused on realistic modeling of plant ecosystems. Starting with a terrain specification and environmental characteristics such as soil humidity, the researchers' virtual plant engine distributed plant species across the terrain. With the engine running on a supercomputer array, a computer interface afforded a user the ability to zoom in and zoom out while investigating the component wildflowers that had grown to fill the terrain.
Once procedural plant models are driven by parameters to a virtual plant growth engine, nature can provide additional inspiration in evolving new plants from existing parameter lists. The simulation process mimics nature by repackaging parameter lists into psuedo-genomes that then simulate physical plant genomes. Once the parameters are in a genome format, the forces of evolution can be written algorithmically in the virtual plant application engine to provide a variety of plant offspring. Chris Colby of Boston University classifies observed phenomena of genome evolution into five processes: mutation, recombination, gene flow, genetic drift, and natural selection. All five processes can be simulated in lines of code. A quick mention of each is warranted here.
Mutation occurs when some external process (such as a cosmic ray) interferes with the normal functioning of DNA within a living cell and forces a change in its chemical makeup. The changed structure then can change the mutated structure's function. Mutation usually does more harm than good to the affected entity but in nature, on average, only about 1 x 10-11 mutations occur per base pair of DNA per generation.
Recombination occurs most spectacularly through meiosis during sexual reproduction. During meiosis, the mother provides half the offspring's genetic material and the father provides the other half. So, the next generation's DNA varies quite dramatically from that DNA of the mother or father's individual.
Gene Flow occurs when some process (such as a mosquito drawing blood) transfers genetic material from one species to another and that material makes its way into the DNA of the target species. The process has been documented between species of fruit flies with a parasitic mite as carrier.
Genetic Drift occurs as a purely statistical process. As a gene pool reproduces and survives in an environment where death is caused by events outside its control, chance determines which genes continue to reproduce and which genes die out.
Natural Selection refers to the process where a gene pool's makeup changes based on reproducing and surviving in an environment where the individual does have some control over their survival. Those individuals with genes that are better suited to the environment tend to reproduce more often than those less suited. Volumes have been written about the sub processes of natural selection: ability to feed, ability to avoid predators, sexual selection, etc. as Charles Darwin intrigued the world with his study of life's struggle in the Galapagos Islands of South America.
All five processes of genetic evolution can be added to a biotic evolution simulation. To demonstrate the genetic processes, we decompose the L-system into components that can be controlled by genetic representation. Take a look at a simple example of an L-system. The model presented on the left is created by the L-system described on the right.


L-Systems incorporate recursion to grow plant structure iteratively. For each recursive processing of the L-system rules, the output of a previous process run is fed as input to the next. The L-system includes rules to apply at each iteration that defines the process by which petals and leaves are generated. As a result, the leaves and petals are similar with the difference being that leaves are created in earlier iterations and are colored green, while petals are created in later iterations and are brightly colored at the red and violet ends of the color spectrum (not that virtual plants have to conform to the stereotypes of physical plants). Alternatively, the petals and leaves can be generated by separate L-system instructions.
L-system examples are available from many different sources. You can learn from exploring each approach to generating plant structure. If you do, as I have done in the past, you will gain insights into how each L-system instruction (in the representation above, each instruction is also called a rule) contributes to overall growth. Eventually, you can pick and choose the best generation techniques and tie them to a genetic representation that identifies which L-system rules to activate for each gene (or group of genes). Through an iterative process of trial and error, my choice for a simulated plant genome became structured to contain a plant expression like the following:
YY|NN|3|0.3 0.3 0.0|0.5|YY NN YN NY|2|4|YY NN YN NY|3|0.2 0.6 0.15|10|1|YY NN YN NY|4|0.6 0.2 0.1
1 2 3 4 5 6 7 8 9 A B C D E F G
Each of the values in the plant genome expands into a combination of 0's and 1's that the genetic simulation uses to perform the five genetic processes described earlier (to be presented as examples below). Using the blue index under each genome grouping as a guide, the components of the genome (left to right) affect the following aspects of growth:
1 - Flowering or not
2 - Leaves on stem or not
3 - Number of stalks
4 - Color of stalk
5 - Effect of gravity on stalk
6 - Shape of stalk
7 - Length of stalk
8 - Number of leaves
9 - Shape of leaves
A - Size of leaves
B - Color of leaves
C - Number of petals
D - Layers of petals
E - Shape of petals
F - Size of petals
G - Color of petals
Once each plant is described by a genome, genetic processes can be applied to future generations of plants by using existing plants as a starting point. Examples of each genetic process using the plant genome above follow.
To simulate mutation, any bit in the value of the genome can be flipped from 0 to 1 or 1 to 0. Although in nature, mutation is rare, simulated mutation can be applied more often to increase the rate of evolution. Referring to the genome index, the right-most feature G represents the color of the petals generated when growing the plant specified by the genome. If we start with a plant that has a red component color that is orange in hue and mutate two bits in that representation (the fourth and sixth of the eight bits), the next generation plant based on that genome produces a saturated red petal instead. Mutation has changed the expression of petal color from an orange-red to a more pure red.

To simulate recombination, a second parent is added from which the next generation is created through meiosis. In the case of petal color, each bit of the resultant genome can come from a parent genome with equal probability. For example, if bits 1, 2, 3, 8 come from a brown petal parent and 4, 5, 6, and 7 come from a red petal parent, the same orange-red petal couple be expressed by the offspring. Alternately, meiosis can occur only along allele boundaries. In that case one allele comes from each parent and the resultant offspring receives its expression from the rules of dominance and recession as seen below. The dominant trait is expressed in every condition except when both parent genomes provide a recessive allele. In the case below which comes from the early part of the genome indexed by the letter B in the figure above, leaves on the stem is a dominant trait so leaves are expressed in the offspring unless an N is provided by each parent (such as the NN expression presented in the L-system rule generation image).
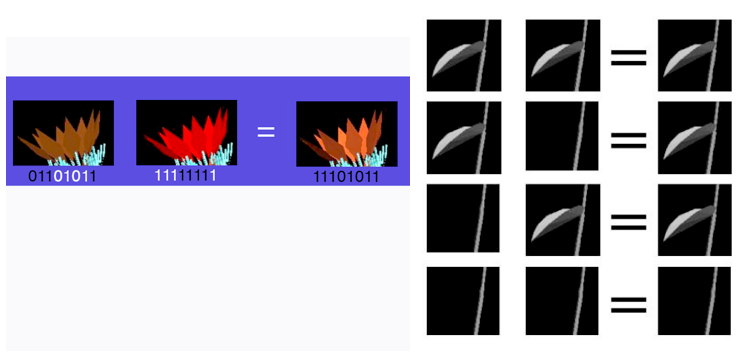
To simulate gene flow, any representative sequence of 0s and 1s can be inserted within the genome to override the previous generation's genome. The simulation of an actual parasite genome sequence known to affect plant genomes to produce representative results helps validate the occurrence of gene flow in the wild. Other biological processes make specific gene flow events beneficial to the host, parasite, or both — more often beneficial to the parasite at the detriment of the host.
Genetic drift can be simulated through adding biases to the plant genomes that get to express themselves in future generations. Through simulating a honey bee that has generated a preference for wavelengths of light in the yellow range of the color spectrum (perhaps from genetic drift through a series of random events), genomes that express yellow plant parts can be expressed in processes that generate the next generation of plants more often than those that express other colors instead. Eventually, through probabilities of genetic processes, the population of plant genomes will drift towards yellow expression on the whole.
Natural selection is perhaps the most interesting genetic process to simulate. To simulate natural selection, an environment is added to the simulation that provides evolutionary pressure on the plants to survive and then thrive. Examples of disease, severe weather, animal and other plant interaction, and long-term climate environmental factors contribute to simulated environments that simulate natural selection pressures on plant genomes. To simulate natural selection, other processes besides plant genome expression need to be included in the simulation — often making the simulation significantly more complex than what the other genetic processes require to be included.
When I connected my plant evolution software to an interactive visualization toolkit called the ARToolkit (augmented reality toolkit), I was able to expose virtual plants to environmental pressures from humans. I seeded a catalog of virtual plants that expressed genomes across the range of possible expressions. I brought the catalog to an art show that lasted fifty-five hours and let the show's attendees mate plants from the catalog and then save their plants for others to use should they desire to do so. During the fifty-five hours, we created just over 11,000 plants as progeny of those original twelve plants in the catalog. There were many environmental pressures provided by the humans, which bunched the resultant offspring into categories of genome expression. For example, the gravity response component to the genome (index 5 in the presentation of the genome above) had certain ranges where attendees thought the plants looked more attractive and interesting than other ranges. As a result they preferred certain parents to others for producing the next generation of plants.
One particular lesson younger attendees learned came from the fact they were impatient when choosing the plants to mate — more interested in seeing the mating occur than anticipating the outcome and planning their choices accordingly. If they mated the same plants over and over from the same two initial parents, they ran out of genetic diversity in the genome and found the next generation looked too much like the previous for their tastes. When they declared that "it's not working anymore", I had a teachable moment to explain how in-breading works and why it is considered unhealthy for a species to do so. The in-breading outcome made sense but I hadn't expected it as an outcome. To get a sense of the process by which attendees mated plants, consider the picture here:

The attendee on the left held a pair of mounted displays that provided images of the flowers draped over a camera image of the physical world. Other attendees could see what she saw through a projection on the far wall in the art show headquarters. The attendee had chosen a plant from previous plants created by attendees (it looked like a relative of an iris) and was moving it within proximity of a plant from the plant catalog (which looked like a relative of a poinsettia). Within five seconds of doing so, a plant mating animation sequence appeared and a new offspring plant was created for everyone's consideration. Others who came to try out the simulation after this attendee's session then had the offspring plant available for their use should they so choose.
Attendees rewarded my contribution of evolutionary plant mating software by offering me insights they had gained when watching the software in action (whether they were driving the plant mating decisions or not). I was bombarded with thousands of interesting comments from artists and non-artists in a general public setting. At least twenty comments were of the quality that I made copious notes so I could consider adding their ideas to the software. The insights came from thoughts inside and outside the fields of botany and evolution — some that showed a deep understanding of botany and evolution and some that showed very little understanding but engaged the attendee to ask questions and seek explanations that deepened their understanding.
L-Systems are but one of many possible systems for expressing genomes in ways that simulations can open up the wonders of genetics and evolution to us humans who are a by-product of those concepts in action over a long period of time. Since simulated evolution can speed up time to produce thousands of generations in the course of a few hours, insights as to possible effects over billions of years can start to be fathomed. Understanding about how genomes hold information for use during the development of living entities unfolds through the simulation of such process. Visual interfaces for watching the software in action through controlling the time and environment in which plant evolutionary process takes place can provide the tinkerer the opportunity to linger whenever insights come and continue when insights are not forth coming.
Exploring plant evolutionary process provides insight into evolutionary process in general. The genetic algorithm, being loosely based on evolutionary theory to include the genetic processes mentioned in this chapter, is being applied to many domains to find optimal solutions to complex decisions that can be evaluated according to agreed upon metrics. Some of those problems don't appear related to any natural evolutionary process, but are structured as virtual evolutionary processes to take advantage of the genetic algorithm. By adding a human in the loop as a decision-maker — whether designing genome expression by hand or choosing which genome expressions are used in generating future generations — we can find sweet spots of interaction between humans and software that provide satisfying results. Results can include a more attractive piece of art or a more energy-efficient airplane wing that is as structurally sound as less energy-efficient alternatives.
If pursued with imagination and creativity, the genetic algorithm can be considered for inclusion in all kinds of simulation software. Certainly the case studies of hydrology and emergency response can use the genetic algorithm to provide environmental factors to which hydrology and emergency response simulations respond. Honing those genomes through evaluation metrics can help validate the realism of simulated results for specific cases that are anticipated as plausible or as good educational examples. The L-System growth component in this chapter is just one particular use of genomes in software — provided because it allows for animating growth iterations of which the number can be set based on the capabilities of the hardware available for running the software. Next Chapter